ML
SVM
https://docs.opencv.org/4.5.0/d1/d73/tutorial_introduction_to_svm.html (opens in a new tab)
https://learnopencv.com/support-vector-machines-svm/ (opens in a new tab)
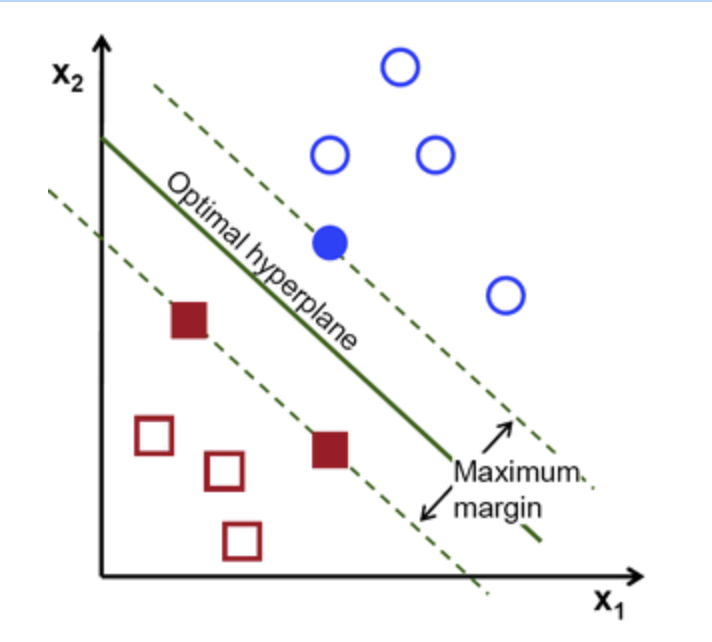
-
Definition of a hyperplane:
-
Canonical hyperplane: by convention, among all the possible, we choose , where represent the training image closest to the hyperplane
-
Distance between a point and hyperplane:
-
The margin M is twice the distance to the closest example:
-
Finally, maximizing M is minimizing with some constraints to correctly classify all training examples
subject to
this is a Lagrangian optimization problem
-
full script
labels = np.array([1, -1, -1, -1]) trainingData = np.matrix([[501, 10], [255, 10], [501, 255], [10, 501]], dtype=np.float32) # train svm = cv.ml.SVM_create() svm.setType(cv.ml.SVM_C_SVC) svm.setKernel(cv.ml.SVM_LINEAR) svm.setTermCriteria((cv.TERM_CRITERIA_MAX_ITER, 100, 1e-6)) svm.train(trainingData, cv.ml.ROW_SAMPLE, labels) # Data for visual representation width = 512 height = 512 image = np.zeros((height, width, 3), dtype=np.uint8) green = (0,255,0) blue = (255,0,0) for i in range(height): for j in range(width): sampleMat = np.matrix([[j,i]], dtype=np.float32) response = svm.predict(sampleMat)[1] if response == 1: image[i,j] = green elif response == -1: image[i,j] = blue thickness = 2 sv = svm.getUncompressedSupportVectors() for i in range(sv.shape[0]): cv.circle(image, (sv[i,0], sv[i,1]), 6, (128, 128, 128), thickness)

-
Non Linear SVM
-
The training data can be rarely separated using an hyperplane.
-
The training data can be rarely separated using an hyperplane
-
Chance is more for a non-linear separable data in lower-dimensional space to become linear separable in higher-dimensional space.
- In general, it is possible to map points in a d-dimensional space to some D-dimensional space to check the possibility of linear separability
-
The new model has to include both the old requirement of finding the hyperplane that gives the biggest margin and the new one of generalizing the training data correctly by not allowing too many classification errors.
-
For example, one could think of minimizing the same quantity plus a constant times the number of misclassification errors in the training data
-
A better solution will take into account the distance of the misclassified samples to their correct decision regions
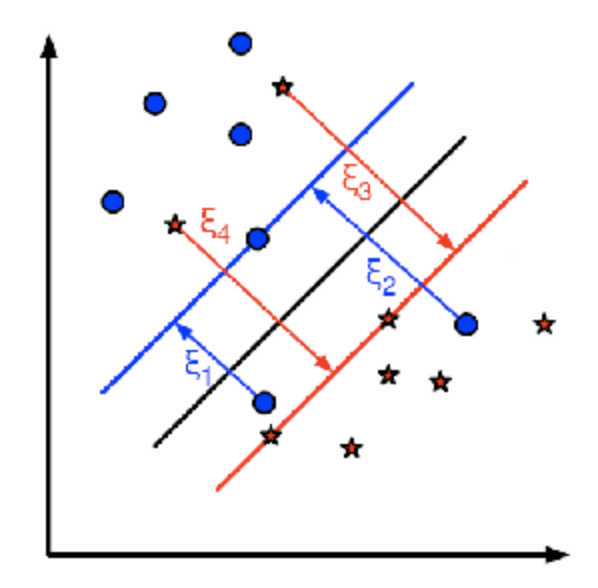
subject to
and
-
-
Choose C
- Large values give solutions with less classification errors but smaller margin
- Small values of C give solutions focusing more on the hyperplane, without much importance for classification errors, so wider margin
-
Implementation
svm = cv.ml.SVM_create() svm.setType(cv.ml.SVM_C_SVC) svm.setC(0.1) # new svm.setKernel(cv.ml.SVM_LINEAR) svm.setTermCriteria((cv.TERM_CRITERIA_MAX_ITER, int(1e7), 1e-6))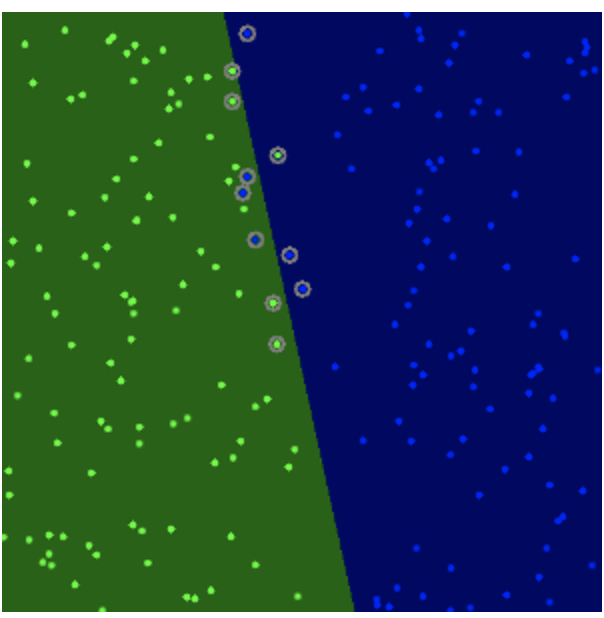
PCA
https://docs.opencv.org/4.5.0/d1/dee/tutorial_introduction_to_pca.html (opens in a new tab)
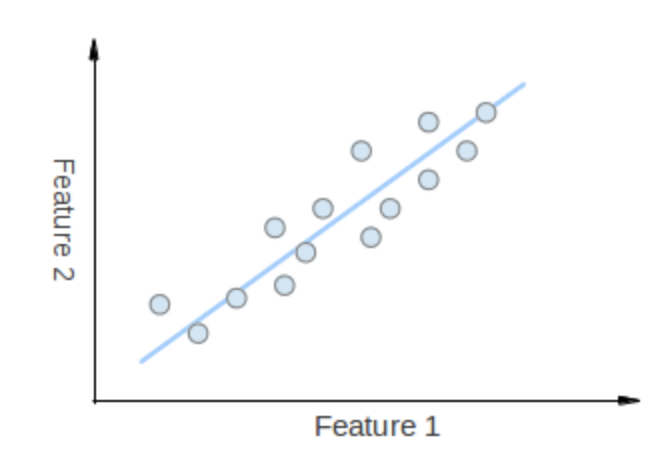
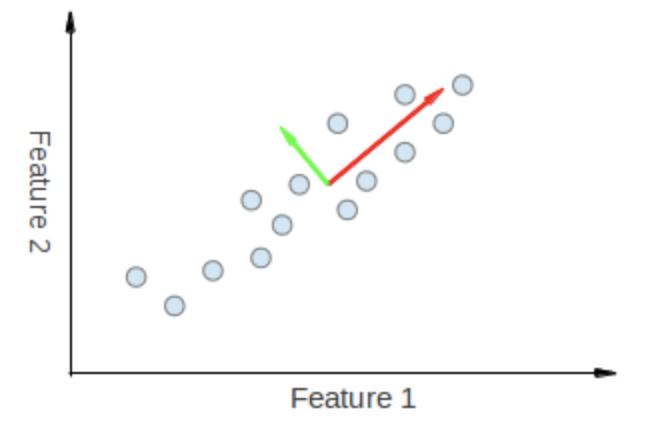
eigenvectors
- The points vary the most along the blue line, more than they vary along the Feature 1 or Feature 2 axes, so we'll be better off with that dimension only
- We want to turn dataset into a small dimension one
- Karhunen-Loève transform:
- Algorithm
-
Compute empirical mean for every dimension
, with vector of size (p, 1)
-
Compute deviation from mean
with , of size (n, 1)
-
Find the covariant matrix of size (p, p)
, where is the conjugate transpose of
If B consists only of real number, it is just the regular transpose.
-
SVD decomposition of the covariant matrix to find eigenvalues and eigenvectors
With , with being the i-th eigenvalues of C
are eigenvectors, come in pairs with eigenvalues
-
full script
from __future__ import print_function from __future__ import division import cv2 as cv import numpy as np import argparse from math import atan2, cos, sin, sqrt, pi def drawAxis(img, p_, q_, colour, scale): p = list(p_) q = list(q_) angle = atan2(p[1] - q[1], p[0] - q[0]) # angle in radians hypotenuse = sqrt((p[1] - q[1]) * (p[1] - q[1]) + (p[0] - q[0]) * (p[0] - q[0])) # Here we lengthen the arrow by a factor of scale q[0] = p[0] - scale * hypotenuse * cos(angle) q[1] = p[1] - scale * hypotenuse * sin(angle) cv.line(img, (int(p[0]), int(p[1])), (int(q[0]), int(q[1])), colour, 1, cv.LINE_AA) # create the arrow hooks p[0] = q[0] + 9 * cos(angle + pi / 4) p[1] = q[1] + 9 * sin(angle + pi / 4) cv.line(img, (int(p[0]), int(p[1])), (int(q[0]), int(q[1])), colour, 1, cv.LINE_AA) p[0] = q[0] + 9 * cos(angle - pi / 4) p[1] = q[1] + 9 * sin(angle - pi / 4) cv.line(img, (int(p[0]), int(p[1])), (int(q[0]), int(q[1])), colour, 1, cv.LINE_AA) def getOrientation(pts, img): sz = len(pts) data_pts = np.empty((sz, 2), dtype=np.float64) for i in range(data_pts.shape[0]): data_pts[i,0] = pts[i,0,0] data_pts[i,1] = pts[i,0,1] # Perform PCA analysis mean = np.empty((0)) mean, eigenvectors, eigenvalues = cv.PCACompute2(data_pts, mean) # Store the center of the object cntr = (int(mean[0,0]), int(mean[0,1])) cv.circle(img, cntr, 3, (255, 0, 255), 2) p1 = (cntr[0] + 0.02 * eigenvectors[0,0] * eigenvalues[0,0], cntr[1] + 0.02 * eigenvectors[0,1] * eigenvalues[0,0]) p2 = (cntr[0] - 0.02 * eigenvectors[1,0] * eigenvalues[1,0], cntr[1] - 0.02 * eigenvectors[1,1] * eigenvalues[1,0]) drawAxis(img, cntr, p1, (0, 255, 0), 1) drawAxis(img, cntr, p2, (255, 255, 0), 5) angle = atan2(eigenvectors[0,1], eigenvectors[0,0]) # orientation in radians return angle parser = argparse.ArgumentParser(description='Code for Introduction to Principal Component Analysis (PCA) tutorial.\ This program demonstrates how to use OpenCV PCA to extract the orientation of an object.') parser.add_argument('--input', help='Path to input image.', default='pca_test1.jpg') args = parser.parse_args() src = cv.imread(cv.samples.findFile(args.input)) # Check if image is loaded successfully if src is None: print('Could not open or find the image: ', args.input) exit(0) cv.imshow('src', src) # Convert image to grayscale gray = cv.cvtColor(src, cv.COLOR_BGR2GRAY) # Convert image to binary _, bw = cv.threshold(gray, 50, 255, cv.THRESH_BINARY | cv.THRESH_OTSU) contours, _ = cv.findContours(bw, cv.RETR_LIST, cv.CHAIN_APPROX_NONE) for i, c in enumerate(contours): # Calculate the area of each contour area = cv.contourArea(c) # Ignore contours that are too small or too large if area < 1e2 or 1e5 < area: continue # Draw each contour only for visualisation purposes cv.drawContours(src, contours, i, (0, 0, 255), 2) # Find the orientation of each shape getOrientation(c, src) cv.imshow('output', src) cv.waitKey()
-
Kmeans
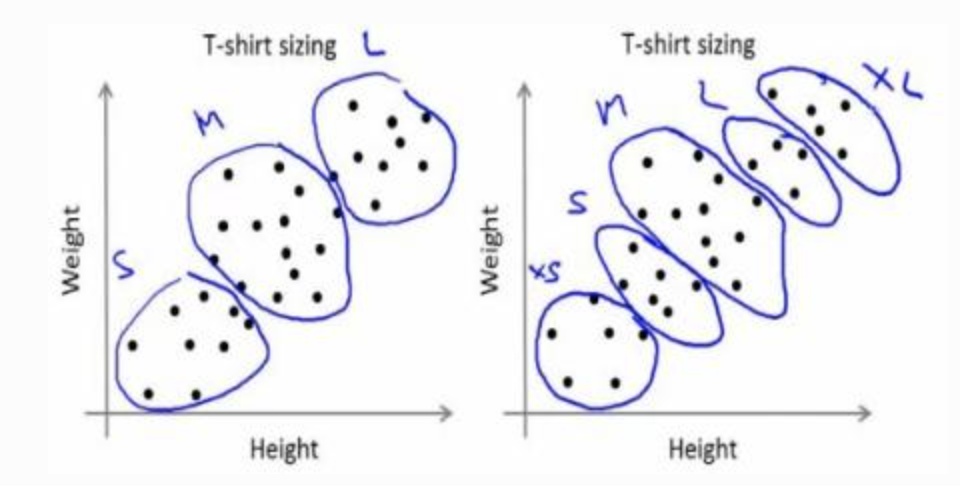
-
Basic intuition
- Randomly choose 2 centroids, and
- Calculate distance for each from both centroid. Closer to are labeled , o.w labeled
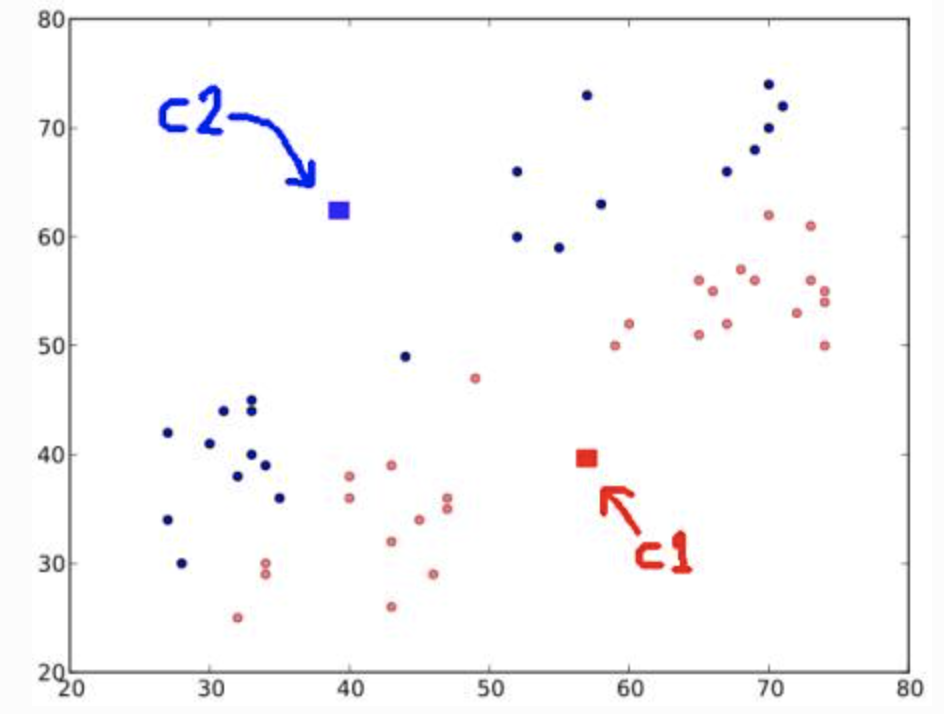
-
Next, compute the average for blue and red points and update the centroids
-
Iterate over 2 and 3 until convergence (or stopping criteria on number of iteration, or precision) is reach
The distances between test data and their centroids are minimum
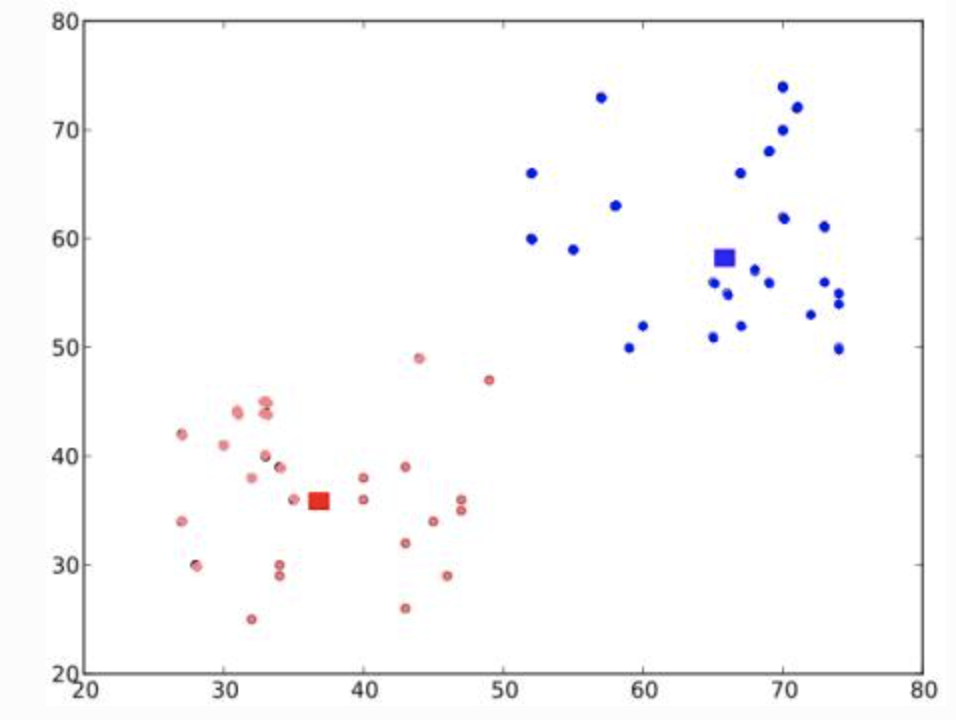
-
full script
import numpy as np import cv2 img = cv2.imread('home.jpg') Z = img.reshape((-1,3)) # convert to np.float32 Z = np.float32(Z) # define criteria, number of clusters(K) and apply kmeans() criteria = (cv2.TERM_CRITERIA_EPS + cv2.TERM_CRITERIA_MAX_ITER, 10, 1.0) K = 8 ret, label, center = cv2.kmeans( Z, K, None, criteria, 10, cv2.KMEANS_RANDOM_CENTERS ) # Now convert back into uint8, and make original image center = np.uint8(center) res = center[label.flatten()] res2 = res.reshape((img.shape)) cv2.imshow('res2',res2) cv2.waitKey(0) cv2.destroyAllWindows()
