9.2 Gaussian Discriminant Analysis
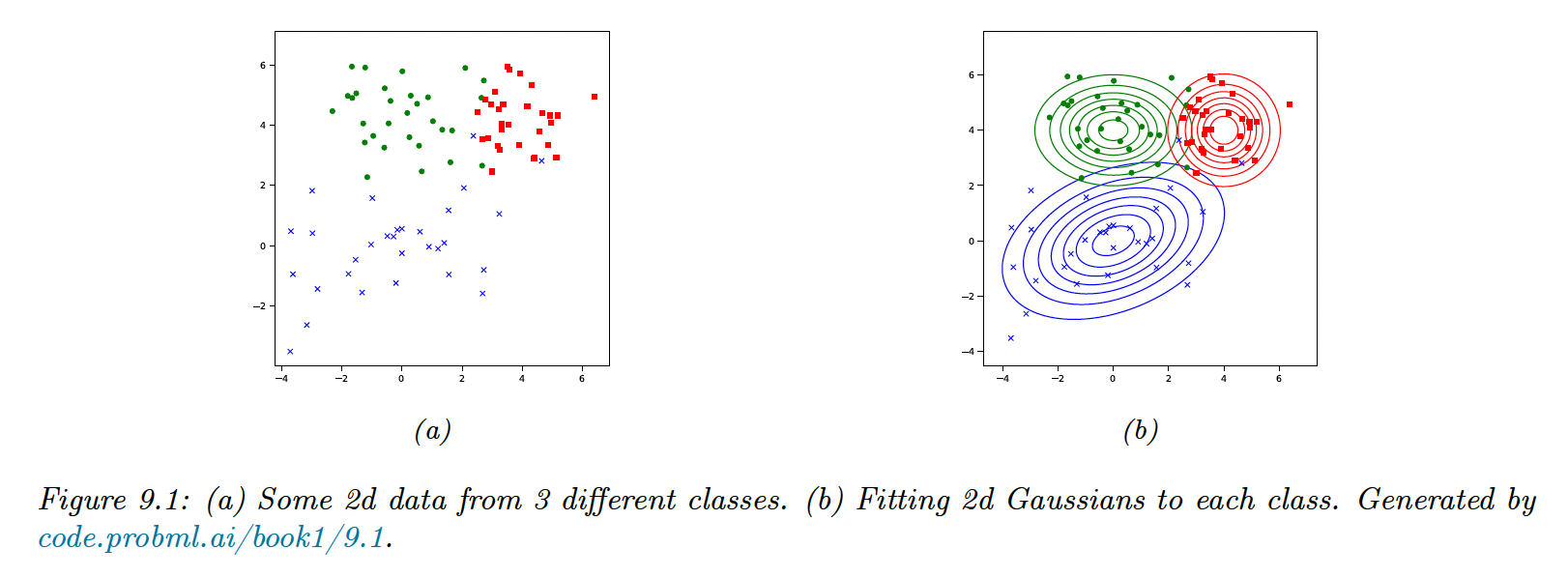
The GDA is a generative classifier where the class conditional densities are multivariate Gaussian:
The corresponding class posterior has the form:
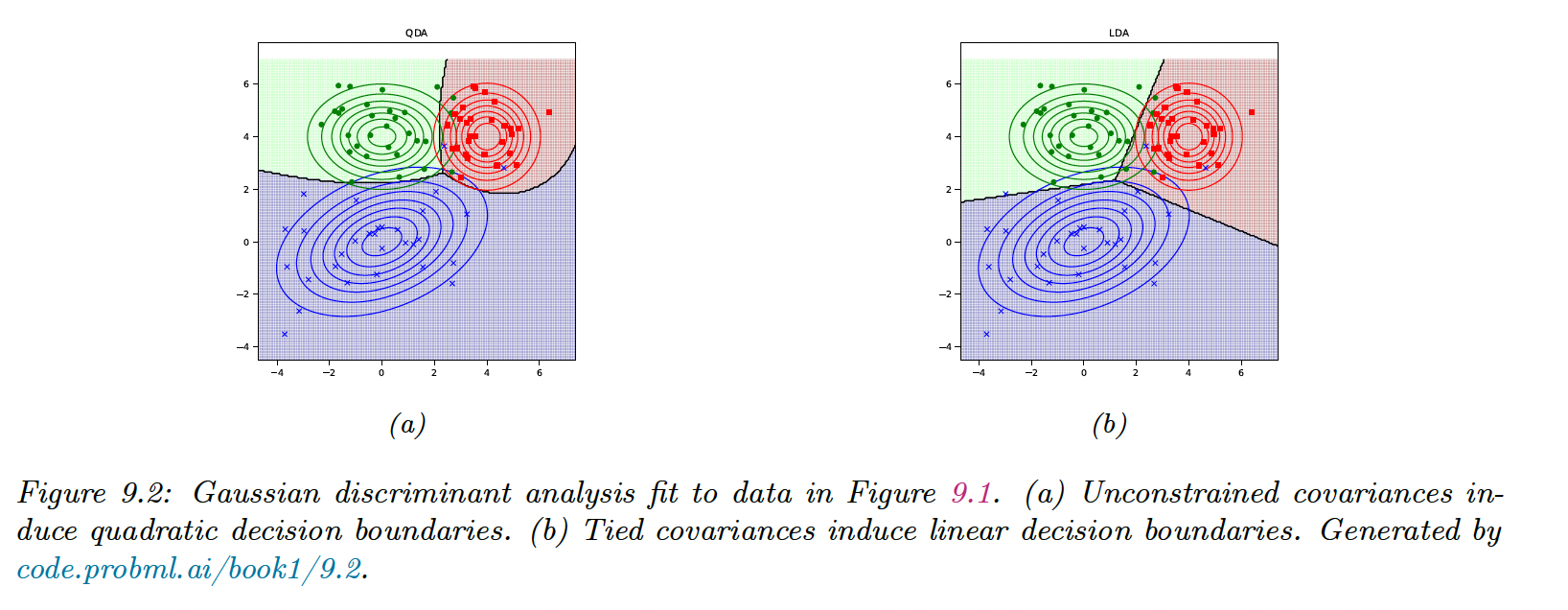
9.2.1 Quadratic decision boundaries
The log posterior over the class labels is:
This is called the discriminant function. The decision function between two classes, , and , is a quadratic function of .
This technique is called Quadratic Discriminant Analysis (QDA).
9.2.2 Linear decision boundaries
We now consider the case of shared covariance, :
is a residual term across classes that can be removed.
This is called the Linear Discriminant Analysis (LDA).
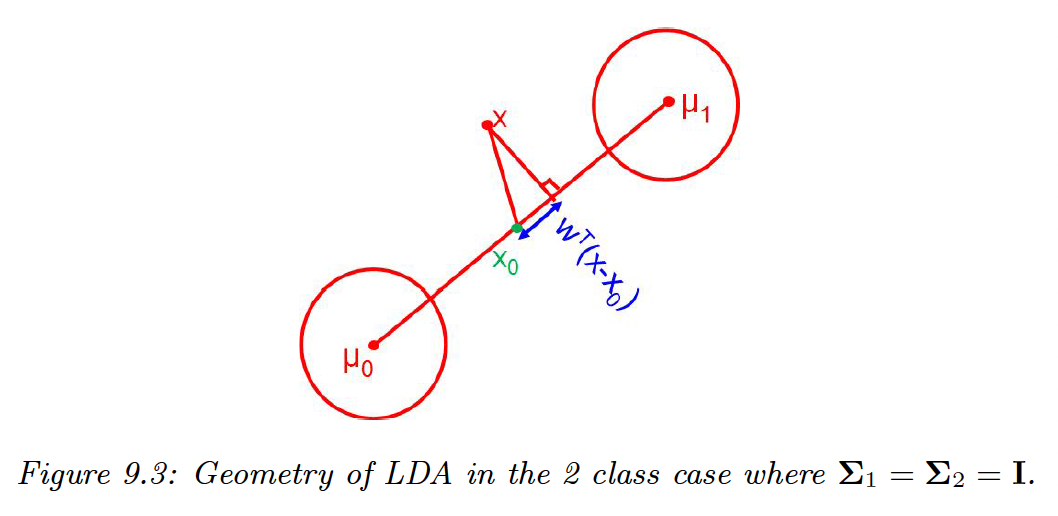
9.2.3 Connection between LDA and Logistic Regression
From the LDA equation, we can write:
- In LDA, we first fit the Gaussians (and class prior) to fit the joint likelihood , then we use to derive
- In Logistic Regression, we estimate directly to maximize the conditional likelihood
In the binary case, this resume to:
With:
And by setting:
We end up with the binary logistic regression:
The MAP decision rule is:
In the case where and , we end up with projecting on the line and compare it to the centroid, .
9.2.4 Model fitting
We now discuss how to fit the GDA model using MLE. The likelihood is:
Hence the log-likelihood is:
We can optimize and separately:
- From section 4.2.4, we have that the MLE of the prior is
- From section 4.2.6, the MLE for the Gaussian is:
Unfortunately, the MLE for can easily overfit if . Below are some workarounds.
9.2.4.1 Tied covariance
If we tied covariance matrices , we get linear boundaries. This also allows pooling from all samples:
9.2.4.2 Diagonal covariance
If we force to be diagonal, we reduce the number of parameters from to , which avoids the overfitting problem. However, we lose the ability to capture the correlation between features (this is the Naive Bayes assumption).
We can restrict further the model using a tied diagonal matrix: this is the diagonal LDA.
9.2.4.3 MAP estimation
Forcing the covariance to be diagonal is a rather strong assumption. Instead, we can perform a MAP estimation from section 4.5.2:
This technique is called regularized discriminant analysis (RDA).
9.2.5 Nearest Centroid Classifier
If we assume a uniform prior over classes, we compute the most probable labels as:
This is called the nearest centroid classifier, where the distance is measured using squared Mahalanobis distance.
We can replace this distance with any other distance metrics:
We can learn distance metrics with one simple approach:
The class posterior becomes:
We can optimize using gradient descent applied to the discriminative loss. This is called nearest class mean metric learning.
This technique can be used for one-shot learning of new classes since we just need to see a single labeled per class (assuming we have a good .
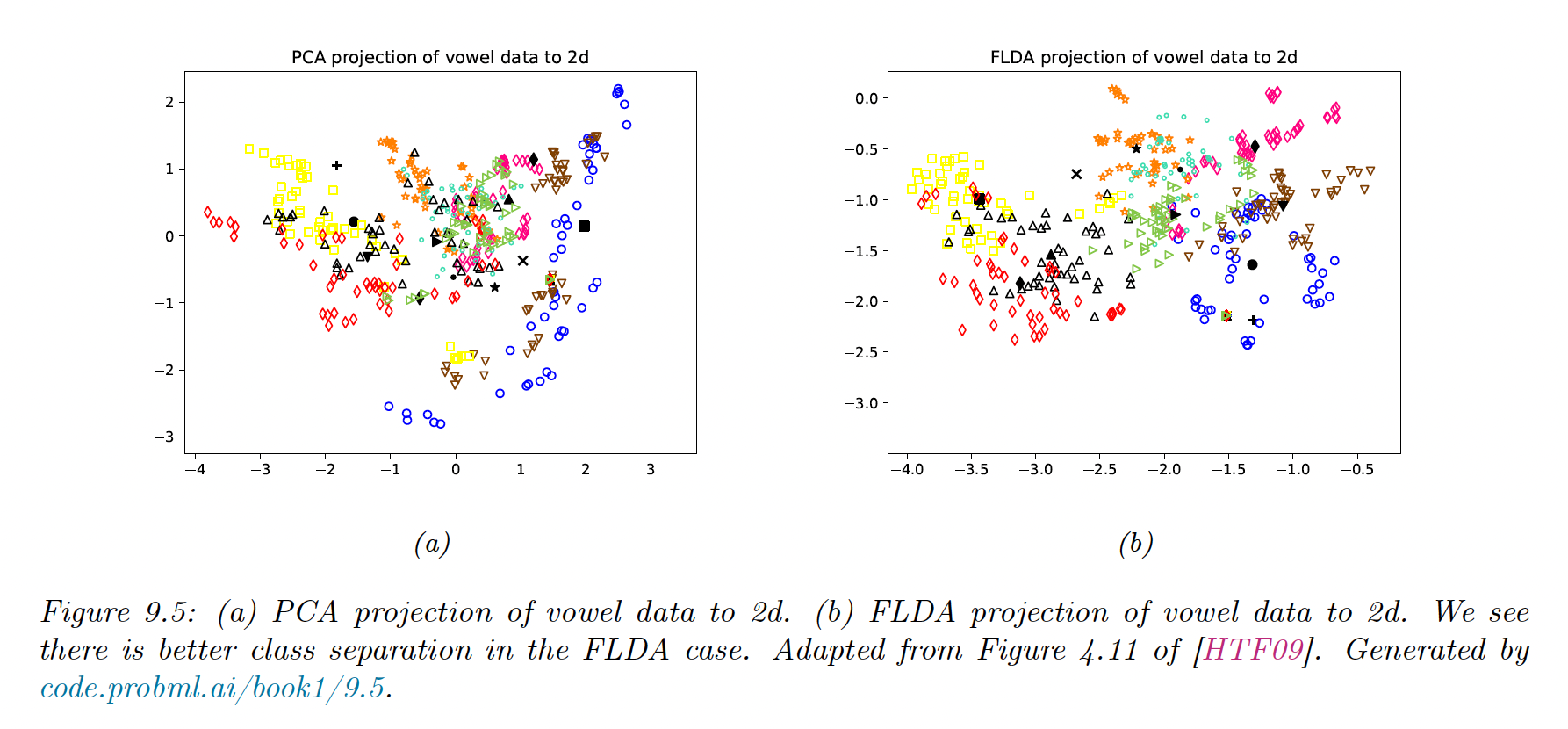
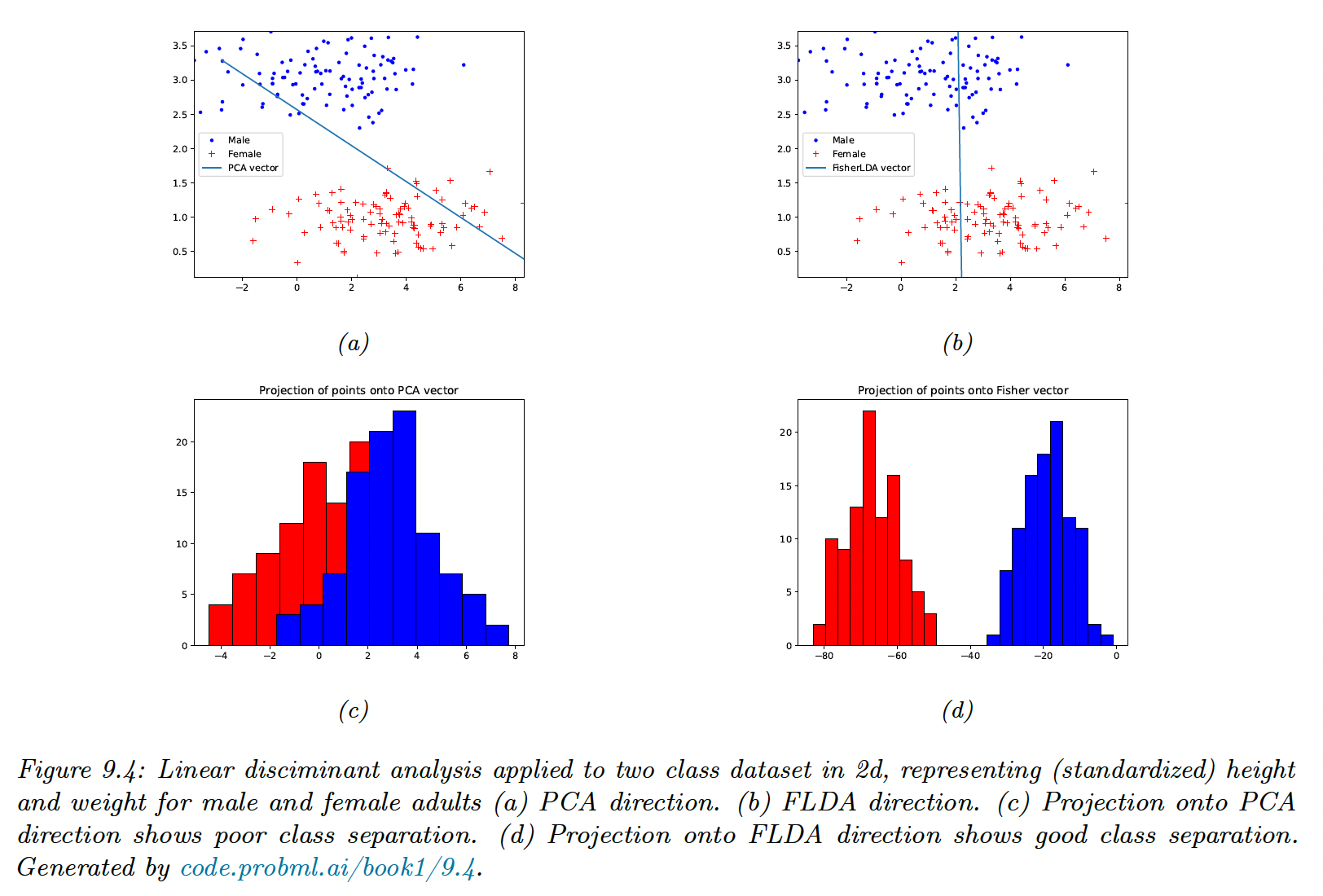
9.2.6 Fisher LDA
Discriminant analysis is a generative approach to classification, that requires fitting MVN to the feature. This might be problematic in high dimensions.
An alternative could be to reduce the features to and then to fit MVN, by finding so that .
- The simplest way is using a PCA, but this is a non-supervised technique, and not taking the labels into account could lead to suboptimal low-dimensional features
- Alternatively, we could use a gradient-based method to optimize the log-likelihood, derived from the posterior in the low dimensional space, as we did with the nearest class mean metric learning
- A third approach relies on eigenvalues decomposition, by finding so that the low dimensional data can be classified as well as possible using a Gaussian class-conditional density model: this is Fisher’s linear discriminant analysis (FLDA)
The drawback of FLDA is that we don’t take into account the dimensionality , since we reduce it to .
In the two classes cases, we are seeking a single vector onto which we project the data.
9.2.6.1 Derivation of the optimal 1d projection
We now estimate the optimal direction . The class-conditional means are:
is the projection of the mean on the line, and is the data projection. The variance of the projected data is:
The goal is to maximize the distance between the conditional means, while minimizing the variance of the projected clusters. We maximize:
where:
- is the between-class scatter matrix
is maximize when:
with
Therefore, if is invertible:
Therefore:
And since we only care about the direction and not the scale:
If , is proportional to , which is intuitive.
9.2.6.2 Multiple classes
We now define:
We have the following scatter matrices:
The objective function is:
This is maximize by:
where are the leading eigenvectors of , assuming non singular.
We see that FLDA discriminates classes better than PCA, but is limited to dimensions, which limits its usefulness.